221 lines
7.6 KiB
Plaintext
221 lines
7.6 KiB
Plaintext
Metadata-Version: 2.1
|
|
Name: dash-cytoscape
|
|
Version: 0.3.0
|
|
Summary: A Component Library for Dash aimed at facilitating network visualization in Python, wrapped around Cytoscape.js
|
|
Home-page: https://dash.plotly.com/cytoscape
|
|
Author: The Plotly Team <cytoscape@plotly.com>
|
|
Author-email: cytoscape@plotly.com
|
|
License: MIT
|
|
Platform: UNKNOWN
|
|
Classifier: Environment :: Web Environment
|
|
Classifier: Framework :: Flask
|
|
Classifier: Intended Audience :: Developers
|
|
Classifier: Intended Audience :: Education
|
|
Classifier: Intended Audience :: Financial and Insurance Industry
|
|
Classifier: Intended Audience :: Healthcare Industry
|
|
Classifier: Intended Audience :: Manufacturing
|
|
Classifier: Intended Audience :: Science/Research
|
|
Classifier: License :: OSI Approved :: MIT License
|
|
Classifier: Programming Language :: Python
|
|
Classifier: Programming Language :: Python :: 3
|
|
Classifier: Programming Language :: Python :: 3.5
|
|
Classifier: Programming Language :: Python :: 3.6
|
|
Classifier: Programming Language :: Python :: 3.7
|
|
Classifier: Topic :: Database :: Front-Ends
|
|
Classifier: Topic :: Scientific/Engineering :: Visualization
|
|
Classifier: Topic :: Software Development :: Libraries :: Application Frameworks
|
|
Description-Content-Type: text/markdown
|
|
Requires-Dist: dash
|
|
|
|
# Dash Cytoscape [](https://github.com/plotly/dash-cytoscape/blob/master/LICENSE) [](https://pypi.org/project/dash-cytoscape/)
|
|
[](https://circleci.com/gh/plotly/dash-cytoscape)
|
|
|
|
A Dash component library for creating interactive and customizable networks in Python, wrapped around [Cytoscape.js](http://js.cytoscape.org/).
|
|
|
|
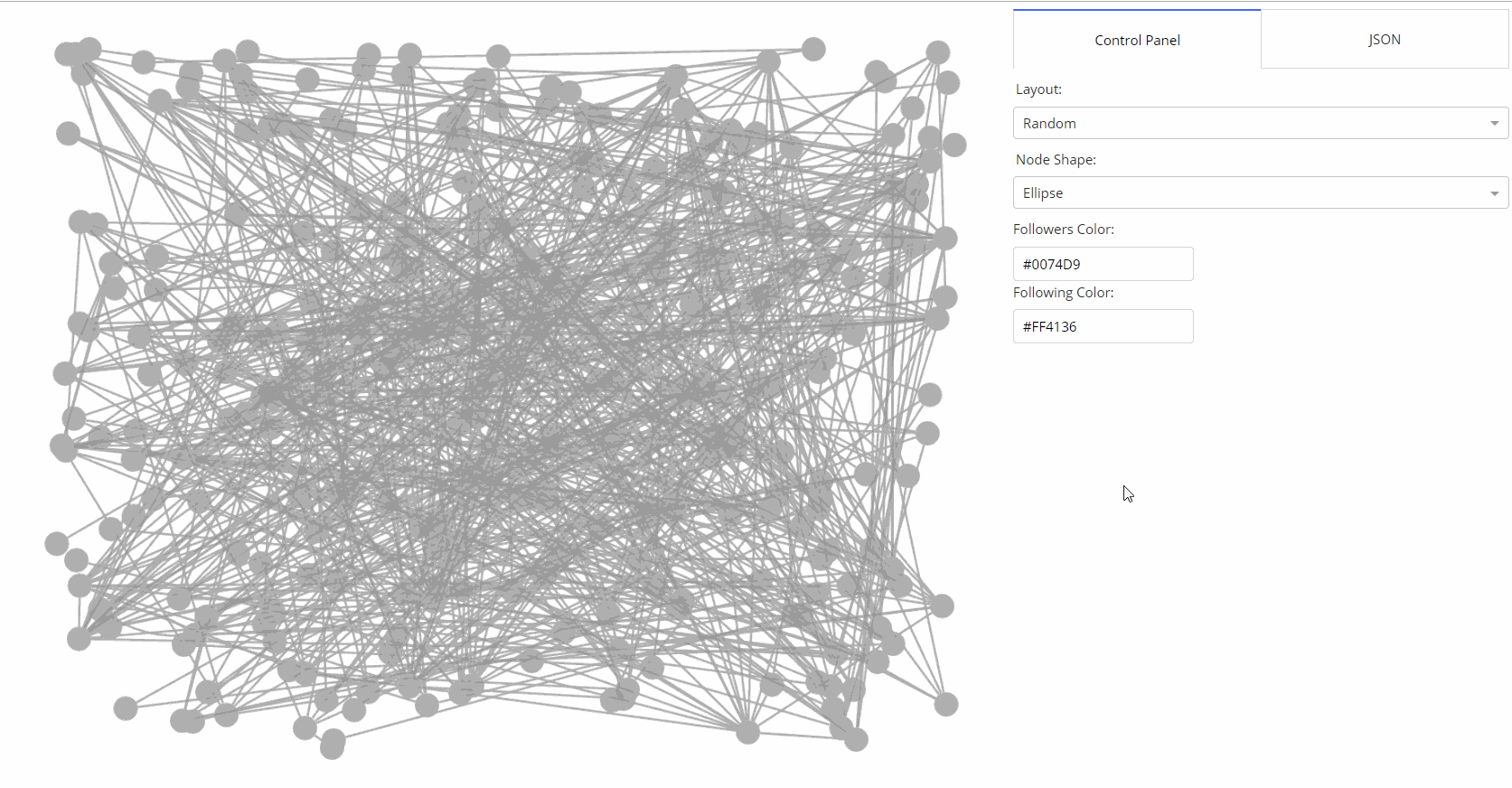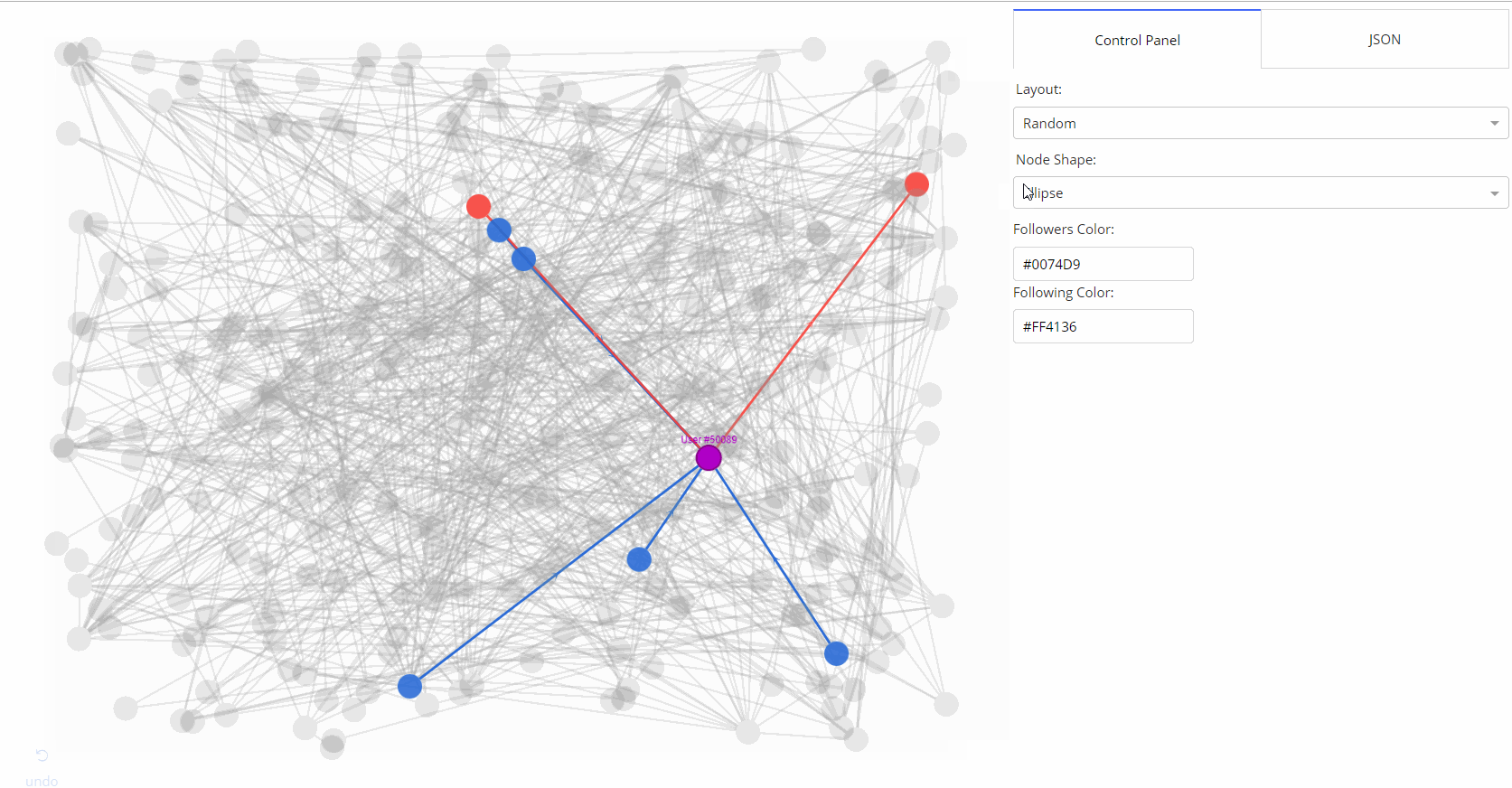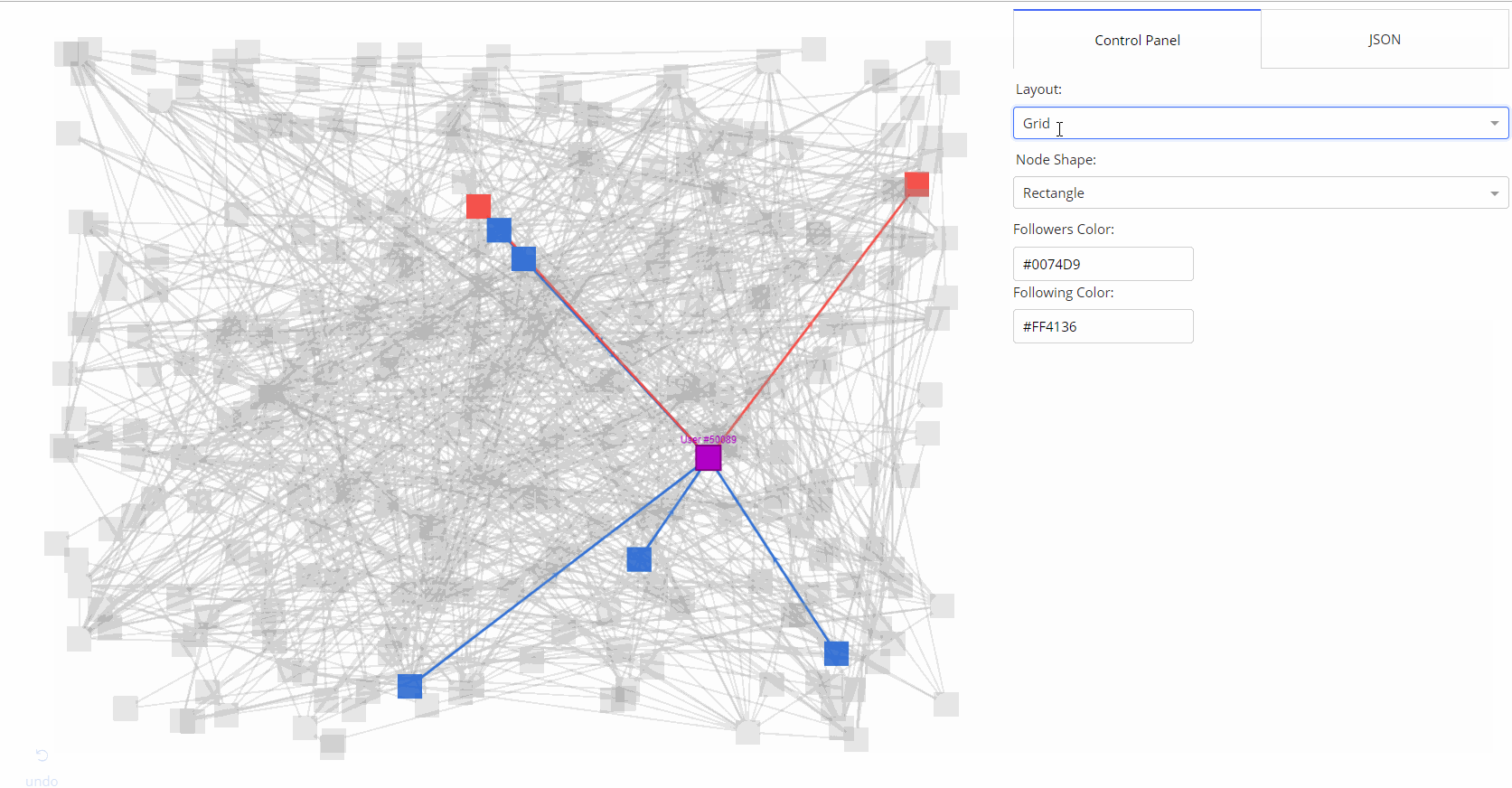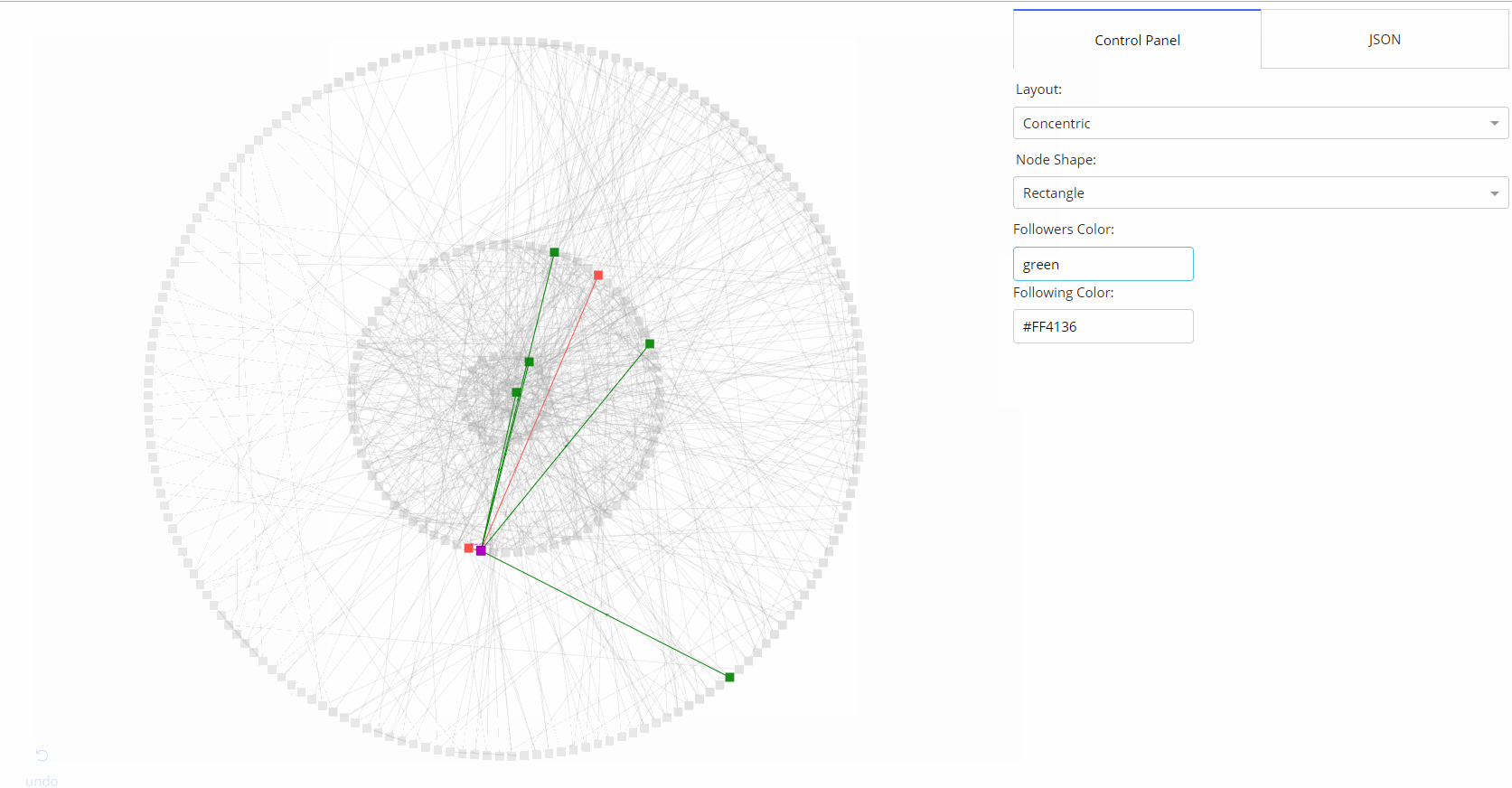
|
|
|
|
* 🌟 [Medium Article](https://medium.com/@plotlygraphs/introducing-dash-cytoscape-ce96cac824e4)
|
|
* 📣 [Community Announcement](https://community.plotly.com/t/announcing-dash-cytoscape/19095)
|
|
* 💻 [Github Repository](https://github.com/plotly/dash-cytoscape)
|
|
* 📚 [User Guide](https://dash.plotly.com/cytoscape)
|
|
* 🗺 [Component Reference](https://dash.plotly.com/cytoscape/reference)
|
|
* 📺 [Webinar Recording](https://www.youtube.com/watch?v=snXcIsCMQgk)
|
|
|
|
## Getting Started in Python
|
|
|
|
### Prerequisites
|
|
|
|
Make sure that dash and its dependent libraries are correctly installed:
|
|
```commandline
|
|
pip install dash
|
|
```
|
|
|
|
If you want to install the latest versions, check out the [Dash docs on installation](https://dash.plotly.com/installation).
|
|
|
|
### Usage
|
|
|
|
Install the library using `pip`:
|
|
|
|
```
|
|
pip install dash-cytoscape
|
|
```
|
|
|
|
Create the following example inside an `app.py` file:
|
|
|
|
```python
|
|
import dash
|
|
import dash_cytoscape as cyto
|
|
import dash_html_components as html
|
|
|
|
app = dash.Dash(__name__)
|
|
app.layout = html.Div([
|
|
cyto.Cytoscape(
|
|
id='cytoscape',
|
|
elements=[
|
|
{'data': {'id': 'one', 'label': 'Node 1'}, 'position': {'x': 50, 'y': 50}},
|
|
{'data': {'id': 'two', 'label': 'Node 2'}, 'position': {'x': 200, 'y': 200}},
|
|
{'data': {'source': 'one', 'target': 'two','label': 'Node 1 to 2'}}
|
|
],
|
|
layout={'name': 'preset'}
|
|
)
|
|
])
|
|
|
|
if __name__ == '__main__':
|
|
app.run_server(debug=True)
|
|
```
|
|
|
|
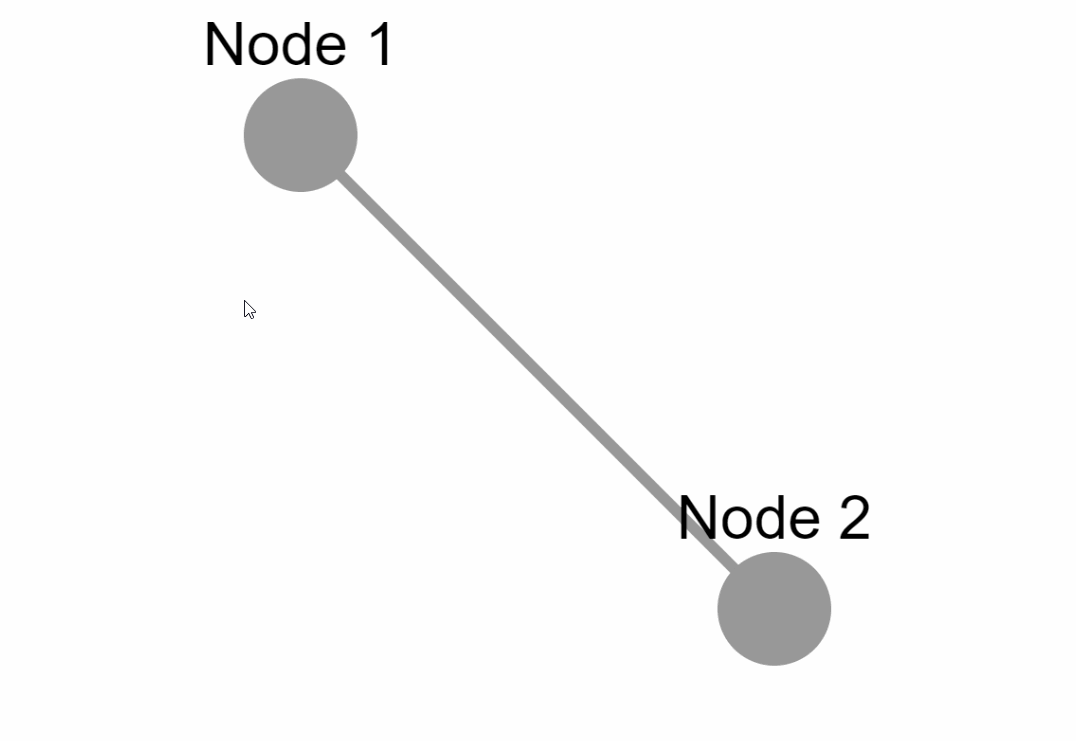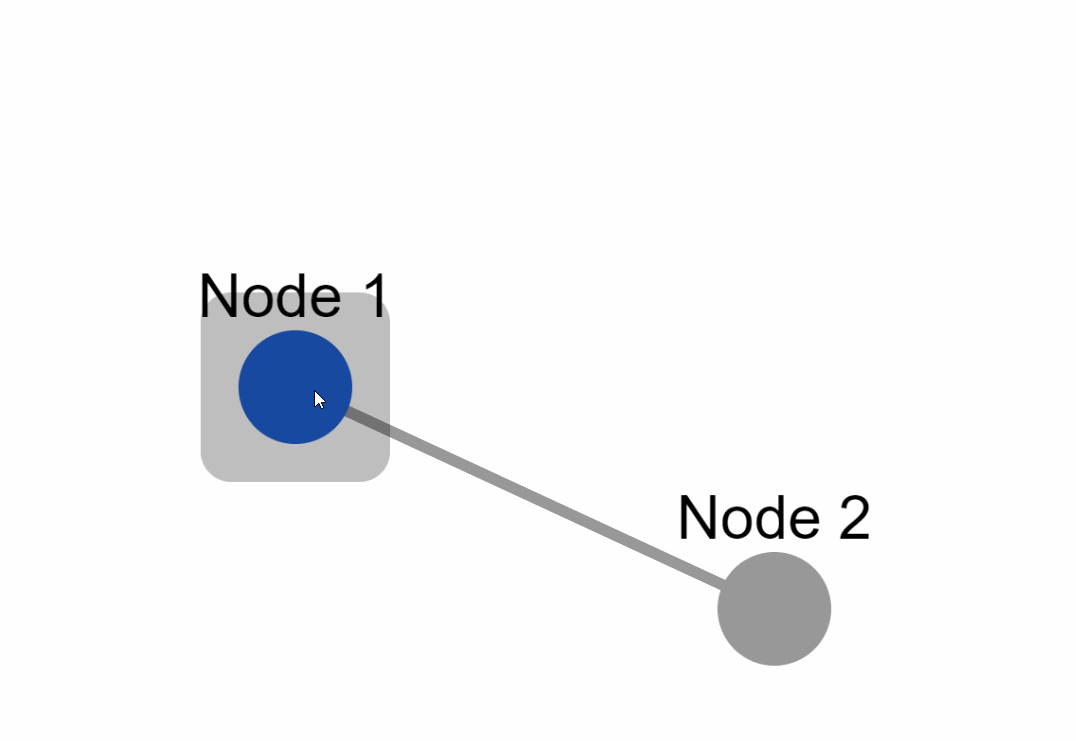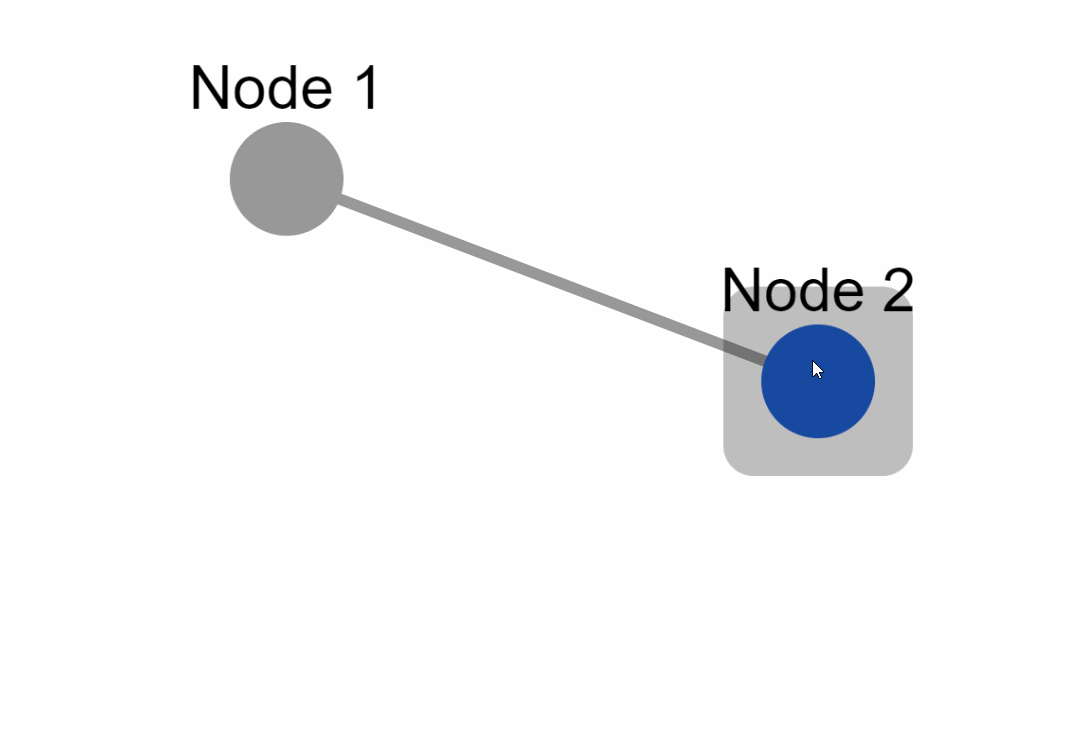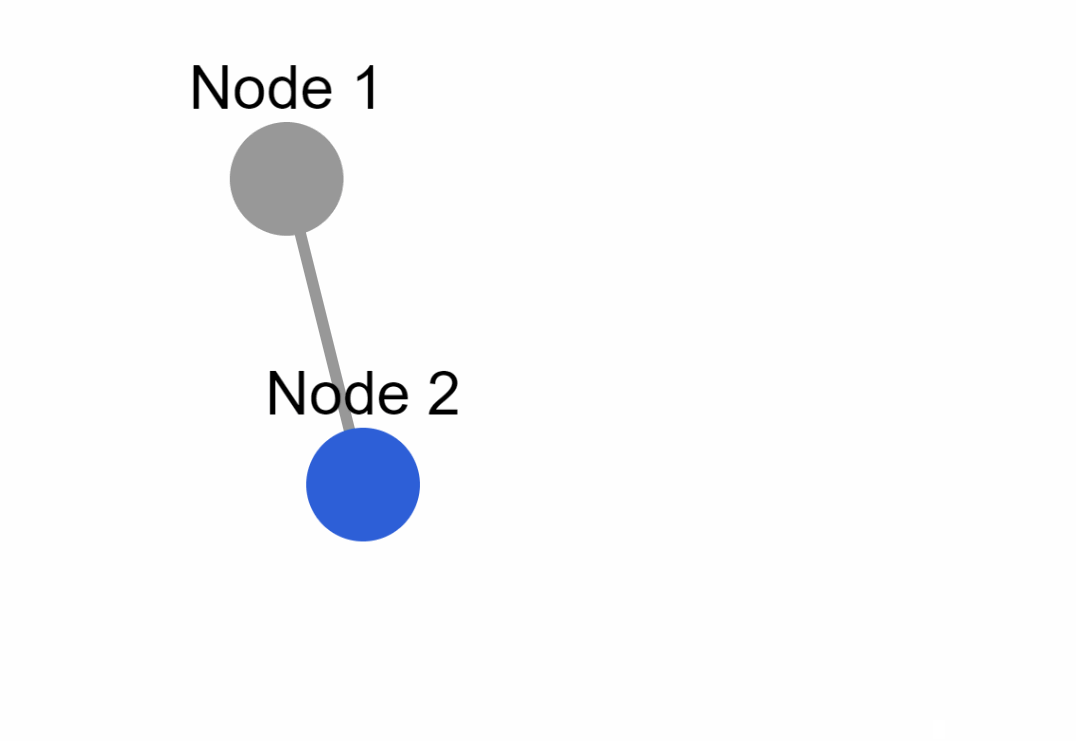
|
|
|
|
### External layouts
|
|
|
|
You can also add external layouts. Use the `cyto.load_extra_layouts()` function to get started:
|
|
|
|
```python
|
|
import dash
|
|
import dash_cytoscape as cyto
|
|
import dash_html_components as html
|
|
|
|
cyto.load_extra_layouts()
|
|
|
|
app = dash.Dash(__name__)
|
|
app.layout = html.Div([
|
|
cyto.Cytoscape(...)
|
|
])
|
|
```
|
|
|
|
Calling `cyto.load_extra_layouts()` also enables generating SVG images.
|
|
|
|
|
|
## Getting Started in R
|
|
|
|
### Prerequisites
|
|
|
|
```R
|
|
install.packages(c("devtools", "dash"))
|
|
```
|
|
### Usage
|
|
|
|
Install the library using devtools:
|
|
|
|
```
|
|
devtools::install_github("plotly/dash-cytoscape")
|
|
```
|
|
|
|
Create the following example inside an `app.R` file:
|
|
|
|
```R
|
|
library(dash)
|
|
library(dashHtmlComponents)
|
|
library(dashCytoscape)
|
|
|
|
app <- Dash$new()
|
|
|
|
app$layout(
|
|
htmlDiv(
|
|
list(
|
|
cytoCytoscape(
|
|
id = 'cytoscape-two-nodes',
|
|
layout = list('name' = 'preset'),
|
|
style = list('width' = '100%', 'height' = '400px'),
|
|
elements = list(
|
|
list('data' = list('id' = 'one', 'label' = 'Node 1'), 'position' = list('x' = 75, 'y' = 75)),
|
|
list('data' = list('id' = 'two', 'label' = 'Node 2'), 'position' = list('x' = 200, 'y' = 200)),
|
|
list('data' = list('source' = 'one', 'target' = 'two'))
|
|
)
|
|
)
|
|
)
|
|
)
|
|
)
|
|
|
|
app$run_server()
|
|
```
|
|
|
|
## Documentation
|
|
|
|
The [Dash Cytoscape User Guide](https://dash.plotly.com/cytoscape/) contains everything you need to know about the library. It contains useful examples, functioning code, and is fully interactive. You can also use the [component reference](https://dash.plotly.com/cytoscape/reference/) for a complete and concise specification of the API.
|
|
|
|
To learn more about the core Dash components and how to use callbacks, view the [Dash documentation](https://dash.plotly.com/).
|
|
|
|
For supplementary information about the underlying Javascript API, view the [Cytoscape.js documentation](http://js.cytoscape.org/).
|
|
|
|
## Contributing
|
|
|
|
Make sure that you have read and understood our [code of conduct](CODE_OF_CONDUCT.md), then head over to [CONTRIBUTING](CONTRIBUTING.md) to get started.
|
|
|
|
### Testing
|
|
|
|
Instructions on how to run [tests](CONTRIBUTING.md#tests) are given in [CONTRIBUTING.md](CONTRIBUTING.md).
|
|
|
|
## License
|
|
|
|
Dash, Cytoscape.js and Dash Cytoscape are licensed under MIT. Please view [LICENSE](LICENSE) for more details.
|
|
|
|
## Contact and Support
|
|
|
|
See https://plotly.com/dash/support for ways to get in touch.
|
|
|
|
## Acknowledgments
|
|
|
|
Huge thanks to the Cytoscape Consortium and the Cytoscape.js team for their contribution in making such a complete API for creating interactive networks. This library would not have been possible without their massive work!
|
|
|
|
The Pull Request and Issue Templates were inspired from the
|
|
[scikit-learn project](https://github.com/scikit-learn/scikit-learn).
|
|
|
|
## Gallery
|
|
|
|
### Dynamically expand elements
|
|
[Code](usage-elements.py) | [Demo](https://dash-gallery.plotly.host/cytoscape-elements)
|
|

|
|
|
|
### Interactively update stylesheet
|
|
[Code](usage-stylesheet.py) | [Demo](https://dash-gallery.plotly.host/cytoscape-stylesheet)
|
|

|
|
|
|
### Automatically generate interactive phylogeny trees
|
|
[Code](demos/usage-phylogeny.py) | [Demo](https://dash-gallery.plotly.host/cytoscape-phylogeny/)
|
|

|
|
|
|
### Create your own stylesheet
|
|
[Code](usage-advanced.py) | [Demo](https://dash-gallery.plotly.host/cytoscape-advanced)
|
|

|
|
|
|
### Use event callbacks
|
|
[Code](usage-events.py) | [Demo](https://dash-gallery.plotly.host/cytoscape-events)
|
|

|
|
|
|
### Use external layouts
|
|
[Code](demos/usage-elements-extra.py)
|
|

|
|
|
|
### Use export graph as image
|
|
[Code](demos/usage-image-export.py)
|
|

|
|
|
|
### Make graph responsive
|
|
[Code](demos/usage-responsive-graph.py)
|
|

|
|
|
|
For an extended gallery, visit the [demos' readme](demos/README.md).
|
|
|
|
|